Lab Rotation Project - I
Prediction of gene expression measurements to understand the origin of complex diseases
systems medicine · genomics · quantitative biology
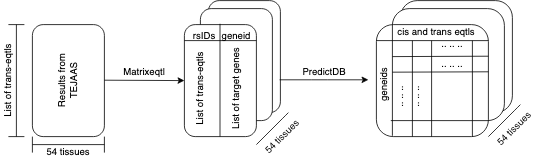
Prediction of gene expression measurements to understand the origin of complex diseases
systems medicine · genomics · quantitative biology

Molecular dynamics simulations of the ribosome - role of ArfB in ribosome rescue
computational biophysics · biomolecular simulations · ribosome complex
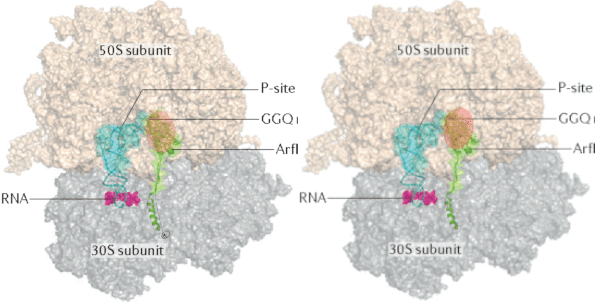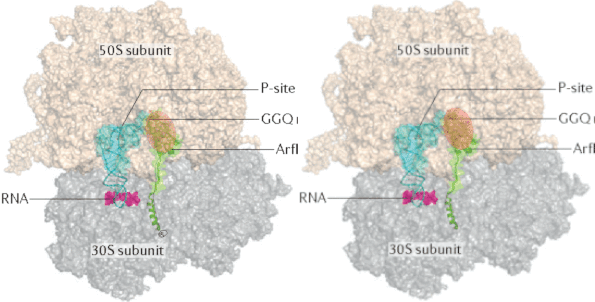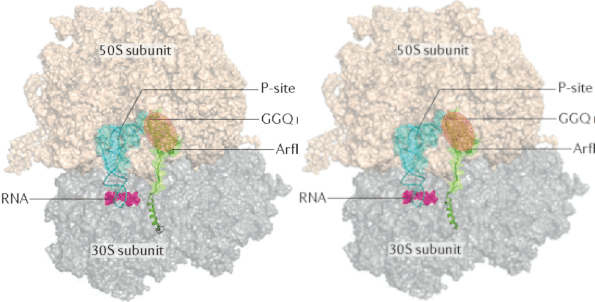
Investigation of ubiquitin dynamics using NMR-based relaxation dispersion measurements
solution state NMR · protein dynamics · mutiscale dynamics

GUI development for a data-based chemical modelling software suite, CANDIY
computational chemistry · protein modelling · protein visualization
Molecular Modelling and Dynamics Studies of Small Molecule Ligands with G-Quadruplex DNA
biomolecular modelling · G4DNA · structural analysis